We have talked about DNA and RNA, now it is time to focus on proteins. Reminder course: DNA is transcribed into RNA which in turn is translated into proteins. These proteins have very diverse roles depending on their tissue and cell location, their ability to transform substrates, their 3D conformation… All the proteins in our body form the proteome and are studied using proteomic techniques.
Proteomics & Aging: How is the proteome studied?
Proteomics is the study of the proteome, which is a set of proteins in a cell. In these analyses, we can look at the changes that a protein undergoes during its synthesis: its translation, post-translational changes (such as acetylation, methylation…), its folding (the 3D structure of a protein allows it to function), its coupling with other proteins (dimer formation, trimers or polymers) or its degradation. Proteins can be found in several forms:
- the primary structure is the succession of amino acids that constitute the protein. It’s the protein sequence. It may be affected by post-translational changes such as methylation or acetylation.
- the secondary structure is the folded form of proteins. This is the structure that the protein takes in order to optimize its future function. Local sheet formation, propellers or bends can be observed.
- the tertiary structure is the 3D form of the protein. Once the secondary structure has been adopted, it must be folded back to give it a conformation in space. This is possible by the formation of bonds between different protein sites.
- the quaternary structure is the final form of proteins, but is not adopted by all proteins. Indeed, it is the result of a combination of several protein chains to form polymers. Some proteins function as monomers and therefore stop at a tertiary structure.
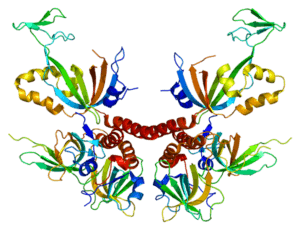
Proteomics makes it possible to study all these structures and identify potential therapeutic targets or biomarkers of pathologies. Its applications are becoming more and more diversified, notably on protein-DNA, protein-RNA and protein-protein interactions, thus joining genomics, transcriptomics and systems biology.
Protein preparation
In order to study the proteome, the different proteins must first be extracted and separated before they are identified and quantified. For this, we start from a tissue (biopsy, organ, blood…) or cells (in culture in particular) that we will destroy to extract proteins. To do this, there are several techniques, the most classic being the use of a chemical product, in combination with anti-proteases (enzymes that degrade proteins and that are blocked), to break cell membranes and recover proteins present inside. This step is extremely important because a good extraction guarantees a good analysis afterwards. It is important to note that, following this extraction stage, proteins are found in their primary form, i.e. without folding or 3D structure. To study secondary and tertiary forms of proteins, there are other techniques, more complex, that we will not detail.
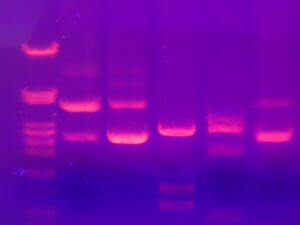
After extraction, we get a protein extract, as pure as possible. It is then necessary to separate the proteins according to their physico-chemical characteristics. The proteins have different sizes, are more or less charged according to their composition, are more or less acidic or basic and will sometimes bind to other proteins called ligands. These properties are therefore used to separate them and electrophoresis or chromatography is most often used.
Electrophoresis consists of a gel placed between two electrodes (one positive and one negative) allowing the passage of a current. This technique will separate the proteins according to their size, the smaller ones migrating faster on the gel, the larger ones remaining in the top of the gel. When a preliminary isoelectric migration step is added, proteins can also be separated according to their charge before being separated by their size. One can thus obtain small spots on a gel, corresponding to proteins.
Chomatography is a completely different technique although it is based on the same properties. The protein extract is injected into a chromatography column (a tube containing filters of various kinds) in order to recover only one type of protein. In the column, we can have size filters, which let only small proteins through, charge filters, which let only positively charged proteins through, for example, or affinity filters, which contain a ligand to which a specific protein will bind. All these different types of columns allow the same type of separation as electrophoresis but have more for purpose the purification of the protein extract than the analysis itself.
Protein identification and quantification
Once proteins have been extracted and purified, proteomics tools allow their identification, analysis and quantification. We can distinguish in particular the western blot technique, which allows proteins to migrate onto an electrophoresis gel, transfer this result onto a membrane and then hybridize this membrane with an antibody specific to our protein of interest. One can thus detect only a given protein, then quantify it thanks to an internal reference to the test. The 2D-DIGE technique (for “Fluorescence two-dimensional differential gel electrophoresis”) is based on the same principle[1]. The main problem with these processes is the limitation to an electrophoresis gel, which does not make large-scale protein analysis possible. Tests allowing the analysis of larger quantities of proteins also exist, such as mass spectrometry or protein chips. The first is quite complex, based on protein labelling using radioisotopes: after digestion by enzymes, several labelled protein fragments are obtained, allowing the quantification and exact identification of all the proteins contained in the sample[2]. Protein chips are very similar to DNA or RNA chips[3].
How to use proteomic data?
The main application of proteomics remains the study of interactions between dysfunctional proteins and pathologies, as well as the development of biomarkers and therapeutic targets of a given disease[4]. Once proteins have been identified and quantified, their abundance (increase or decrease in synthesis) and their form (mutation, modifications, etc.) can be linked to a disorder. Like genomic and transcriptomic data, the results of a proteomic study provide a better understanding of pathologies, at a different and complementary level to DNA and RNA studies[4].
Some examples of protein modifications
Protein sequence modification
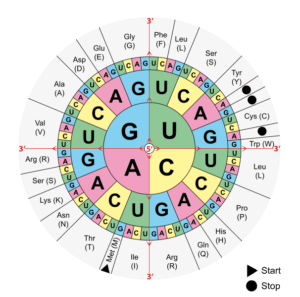
DNA contains information that allows RNA to be translated into proteins according to a specific amino acid sequence. When a mutation occurs in DNA, it will affect the protein sequence. This is called the genetic code: the combination of three DNA bases will give a codon and there are currently 61 different codons giving rise to 20 amino acids. For example, a TTT codon (Thymine x3) will give a phenylalanine (one of the 20 amino acids), while a TCT codon (Thymine-Cytosine-Thymine) will give a serine. This may seem like a small difference, but the final protein structure depends entirely on its amino acid composition. If there is a mutation of a TTT codon to TCT, then phenylalanine will be replaced by a serine in the protein, creating potential problems in terms of folding, active site or binding site.
Alteration of post-translational changes
If the protein has been properly synthesized, it is not immune to other problems. A protein is not limited to its sequence, it must be modified to be functional. These post-translational modifications are very varied and may consist of the addition of phosphate groups (phosphorylation), acetyl groups (acetylation), methyl groups (methylation), various sugars (glycosylation) or ubiquitin groups (ubiquitinylation), the latter being a cell recognition system allowing protein elimination. All these changes can be altered, increased or decreased, affecting the function of the protein.
Modifications of the 3D structure
The final form of a protein consists in its folding in space and its possible association with other proteins. In order to effect its folding, it is necessary to create bonds between the different parts of the protein. These bonds are diverse and can range from strong bonds, such as covalent bonds holding our molecules, to weak bonds, such as ionic bonds, which are based on the interaction between positively charged and negatively charged areas within the protein.
What consequences for our bodies?
Let’s take a simple example known to all: cystic fibrosis. It is a rare genetic disease affecting the mucous membranes and excretion system of our body. The first symptoms are usually pulmonary, with an accumulation of mucus preventing normal breathing, then spreading to the liver, intestine, pancreas or even genital mucosa, causing death between 20 and 40 years.
Why talk about this disease? Because it is the very example of a protein dysfunction. Indeed, the gene coding for the CFTR protein is mutated, resulting in the formation of a truncated protein, unable to fold properly and lodge in the cell membrane as it should be. Unfortunately, there are many other examples of this type of disease whose origin is not necessarily a genetic mutation.
Proteomics applied to aging
It is obvious that beyond their usefulness for rare diseases, proteomic tools also have an interest in research against ageing. Indeed, the major interest of these tests is the large-scale approach and the taking into account of thousands of data chunks processed later thanks to bioinformatics techniques[5]. This allows the identification of new proteins central to the development of pathologies.
For example, a recent study dissected the signaling pathways leading to the accumulation of amyloid β in Alzheimer’s disease, revealing new hypotheses[6]. Another looked at kidney aging in rats, associating 66 proteins with slow aging and 73 with accelerated aging. This study suggests that the renal environment, particularly with respect to oxidative stress and mitochondrial function, is of primary importance in renal aging and degradation of its function[7]. In mitochondria, a team has also developed new tools to track its function over time[8]. Also in mitochondria, recent proteomic studies have identified sites of acetylation, succinylation, glutarylation and malonylation (all post-translational modifications) and their regulation by sirtuins, SIRT3 and SIRT5, proteins largely involved in aging[9]. Concerning age-related sarcopenia, one team related oxidative status to muscle loss[10]. Other studies have examined proteostasis, the loss of which is one of the 9 causes of aging, and its effect on longevity[11].
The list of research in progress could continue over and over again, as the hopes carried by proteomics are important in the fight against aging, but we will focus on these few examples. For more information on the proteome and studies currently under development, you can visit the Human Proteome Project website or the EBI website, to go further in your proteomics training!
See all our articles on the “-omics” approaches
The “-omics” : a tool to better understand our aging process
What is behind the term “-omics? When we talk of genomics, transcriptomics or proteomics, what are we looking at? Here is a guide to explain it all!
Part 1: Understanding genomics for anti-aging research

How can we not mention genomics and how useful they are. It is the oldest “-omics” approach and still the most studied one even today. It gave birth to whole new concepts, such as epigenetics or systems biology, and opened up the scientific community to new horizons that we had never even dreamed ot!
Part 2: Transcriptomics, a constantly evolving science.

The discovery of non coding RNA led to a Nobel Prize. Enough to say it’s an important field of research! Transcriptomics is allowing the discovery of new tools, new mechanisms and led us to better understand the regulation of transcription.
Part 3: Proteomics, a mish-mash of research fields
 Above all, proteomics are a multidisciplinary approach, taking into account the interactions between fields, namely genomics and transcriptomics. It also refers to different concepts, from immunology to nutrition or cell function.
Above all, proteomics are a multidisciplinary approach, taking into account the interactions between fields, namely genomics and transcriptomics. It also refers to different concepts, from immunology to nutrition or cell function.
Part 4: Metabolomics, the last-born of the “omics”
 Last but not least! Metabolomics is a field that helps us understand very complicated regulation networks and the daily discovery of new cell-cell communication players.
Last but not least! Metabolomics is a field that helps us understand very complicated regulation networks and the daily discovery of new cell-cell communication players.
References
[1] R Tonge, J Shaw, B Middleton, et al. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1(3):377–96
[2] RW Nelson, JR Krone, AL Bieber, P Williams, Peter. Mass Spectrometric Immunoassay. Analytical Chemistry. 1995;67(7):1153–1158
[3] DH Wilson, DM Rissin, CW Kan et al. The Simoa HD-1 Analyzer: A Novel Fully Automated Digital Immunoassay Analyzer with Single-Molecule Sensitivity and Multiplexing. J Lab Autom. 2016;21(4):533–47
[4] AD Weston, L Hood. Systems Biology, Proteomics, and the Future of Health Care: Toward Predictive, Preventative, and Personalized Medicine. Journal of Proteome Research. 2004;3(2):179–96
[5] S Rogers, M Girolami, W Kolch, KM Waters, T Liu, B Thrall, HS Wiley. Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics. 2008;24(24):2894–2900
[6] Yu L, Petyuk VA, Gaiteri C, Mostafavi S, et al. Targeted brain proteomics uncover multiple pathways to Alzheimer’s dementia. Ann Neurol. 2018. doi: 10.1002/ana.25266. [Epub ahead of print]
[7] Li D, Zhao D, Zhang W, Ma Q et al. Identification of proteins potentially associated with renal aging in rats. Aging (Albany NY). 2018. doi: 10.18632/aging.101460. [Epub ahead of print]
[8] Cortassa S, Sollott SJ, Aon MA. Computational Modeling of Mitochondrial Function from a Systems Biology Perspective. Methods Mol Biol. 2018;1782:249-265
[9] Carrico C, Meyer JG, He W, Gibson BW, Verdin E. The Mitochondrial Acylome Emerges: Proteomics, Regulation by Sirtuins, and Metabolic and Disease Implications. Cell Metab. 2018;27(3):497-512
[10] Smith NT, Soriano-Arroquia A, Goljanek-Whysall K, Jackson MJ, McDonagh B. Redox responses are preserved across muscle fibres with differential susceptibility to aging. Journal of Proteomics. 2018;177:112-123
[11] Basisty N, Meyer JG, Schilling B. Protein Turnover in Aging and Longevity. Proteomics. 2018 Mar;18(5-6):e1700108
Dr. Marion Tible

Author/Reviewer
Auteure/Relectrice
Marion Tible has a PhD in cellular biology and physiopathology. Formerly a researcher in thematics varying from cardiology to neurodegenerative diseases, she is now part of Long Long Life team and is involved in scientific writing and anti-aging research.
More about the Long Long Life team
Marion Tible est docteur en biologie cellulaire et physiopathologie. Ancienne chercheuse dans des thématiques oscillant de la cardiologie aux maladies neurodégénératives, elle est aujourd’hui impliquée au sein de Long Long Life pour la rédaction scientifique et la recherche contre le vieillissement.
En savoir plus sur l’équipe de Long Long Life
Dr Guilhem Velvé Casquillas

Author/Reviewer
Auteur/Relecteur
Physics PhD, CEO NBIC Valley, CEO Long Long Life, CEO Elvesys Microfluidic Innovation Center
More about the Long Long Life team
Docteur en physique, CEO NBIC Valley, CEO Long Long Life, CEO Elvesys Microfluidic Innovation Center
En savoir plus sur l’équipe de Long Long Life


![[Video] Eurosymposium on Healthy Ageing, Brussels, 2018 Eurosymposium on Healthy Aging](http://www.longlonglife.org/wp-content/uploads/2019/07/P1310252-218x150.jpg)