Aging : Molecular mechanisms of epigenetic alteration
Epigenetics is the study of the mechanisms that act on the genome to control gene expression. Modulating gene expression is important for maintaining cellular balance. Epigenetic alterations lead to changes in gene expression, which can eventually disrupt cellular functions. These changes are recognized as one of the causes of aging and are found in many human diseases.
Controlling gene expression is largely achieved by controlling the condensation state of the chromatin. Highly condensed (heterochromatin), it will not allow gene expression, whereas a poorly condensed chromatin (euchromatin) will. These changes of state are achieved through three mechanisms: DNA methylation, histone modifications and chromatin remodelling. With age, these mechanisms are disrupted, influencing longevity and the development of age-related diseases.
Epigenetics & aging : DNA methylation
DNA methylation aberrations are the most common and studied epigenetic alterations. Over time, methylation abnormalities accumulate and contribute to epigenetic drift during aging. DNA methylation is the addition of a methyl chemical group (-CH3) on a cytosine base (C) of a CpG dinucleotide (a cytosine base followed by a guanine base and linked together by a phosphate bond). Many of these CpG dinucleotides are grouped together in genomic regions called CpG islets (nucleotide sequences with a high concentration of CpG dinucleotides) and about half of these CpG islets are located in gene regulatory regions[1]. It is easy to understand that methylation plays an important role in controlling gene expression.
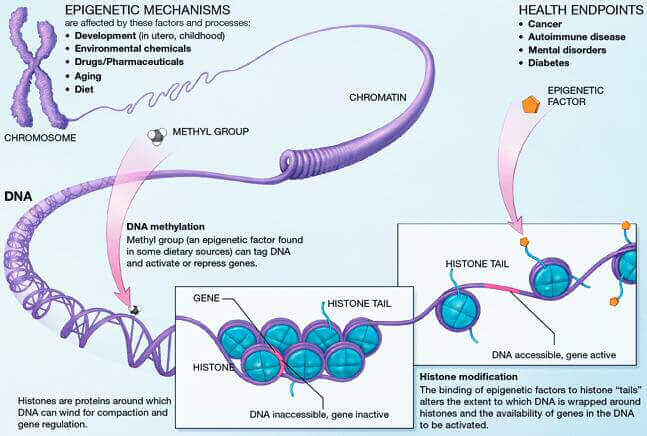
Extensive studies of the methylation levels of CpG islets have shown that there are regions that are differently methylated with age[1]. Some of these specific sites are associated with aging and studying their methylation levels helps to define the biological age of our cells: it is the epigenetic clock. DNA methylation has thus been developed as a powerful and accurate biomarker of biological age prediction[2].
As we age, we observe an overall loss of DNA methylation level throughout the genome (hypomethylation), but some regions are more prone to an increase in methylation level (hypermethylation). DNA methylation is linked to gene repression since it induces the condensation of chromatin, blocking the access of transcriptional machinery to DNA. This repression prevents the expression of oncogenes (genes that promote the development of cancers). But with age-related hypomethylation, these oncogenes can be expressed more abundantly and participate in the development of cancers. Conversely, hypermethylation affects certain regulatory regions of genes that are normally poorly methylated[3,4], this is particularly the case for tumor suppressor genes. Hypermethylation on these genes also increases the risk of developing cancers. Changes in methylation are therefore readily associated with the development of diseases, particularly cancers.
Epigenetics & aging : Histone modifications
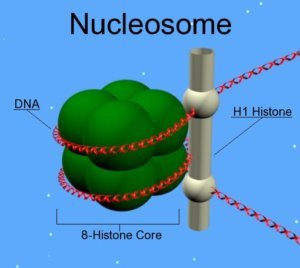
Histones are the constituent proteins of nucleosomes, complexes around which DNA curls up to form chromatin. They undergo modifications that will change the state of compaction of the chromatin, thus controlling gene expression. These post-translational modifications are added or subtracted by enzymes. During aging, the abundance and activity of these enzymes decline, causing alterations in the epigenome. In the long term, this causes the loss of gene balance and promotes the development of age-related diseases[3].
The most common changes affecting histones are acetylations and methylations on lysine residues. Aberrations of these changes have been observed in elderly model organisms and in people with progeria, a rare disease that causes an accelerated aging phenotype[5].
A decrease in overall histone acetylation has been shown to play an important role in mouse brain aging and would result in memory loss[6]. In the same way, a high level of this acetylation is associated with longevity. In addition, methylation changes on H3 histones are associated with the loss of heterochromatin structure, which is recognized as a mark of aging. Histone methylation changes are associated with the development of cancer[4]. In addition to the direct impact of histone methylation and acetylation aberrations on aging, histone methylation also impacts pathways involved in aging, such as autophagy, response to environmental stresses and cell senescence.
Over time, histones undergo other phenomena. There is an overall loss of their numbers in the genome with age. There are also histone variants, crucial for the regulation of chromatin dynamics and found under certain cellular conditions. The H3.3 variant of histone H3 is the form found predominantly in the chromatin of senescent cells, and an excess of this variant in a cell causes cell senescence. Another age-related variant also associated with senescence is the macroH2A variant of histone H2A, whose level increases with age. [6,7]
Epigenetics & aging : chromatine remodeling
In addition to DNA methylation and histone modifications, other molecular mechanisms influence the structure of chromatin through specific proteins. This is the case with HP1 proteins (Heterochromatin protein 1) which contribute to the formation of heterochromatin. As we age, the amount of HP1 protein decreases, which ultimately leads to a decrease in heterochromatin levels, which are recognized as a sign of aging. The loss of heterochromatin that accompanies aging leads to changes in the overall architecture of the nucleus and gene expression, directly or indirectly causing aging and senescence[6].
In particular, HP1 proteins maintain the link between peripheral heterochromatin and the internal nuclear membrane. The loss of this bond due to the decrease in HP1 protein levels leads to chromatin disruption and defective epigenetic regulation. This phenotype is found in patients with Huntchinson-Gilford syndrome (progeria)[8].
Also, the NuRD protein complex (Nucleosome Remodelling Deacetylase) regulates gene expression through two enzymatic activities: nucleosome remodeling through ATP (ATP is the energy of the cell) and deacetylase activity (the acetyl group is removed) on histones. Due to the many subunits that make up the protein complex and their heterogeneity (multiple isoforms), the effects of NuRD are complex and sometimes antagonistic. This is the case in cancers, where depending on the tissue and type of subunit, NuRD will have different incidences on the disease[9]. Current studies will provide a better understanding of its role in other age-related diseases.
Cellular aging can also be accompanied by dysfunction of Polycomb group proteins. It is a group of transcriptional repressors that targets, among other things, genes encoding transcription factors, signaling proteins and regulators of all major developmental pathways. Inappropriate activity of these proteins plays a role in the carcinogenesis, development and progression of cancer[10,11]
Towards epigenetics for aging
Epigenetic alterations have been widely studied since these mechanisms have been clearly defined as one of the causes of aging. At present, entire genomes have been studied to determine the epigenome, i.e. all the epigenetic modifications of the cell. Epigenomics provides an understanding of the impact of epigenetics on aging and opens up research and treatment opportunities to understand the aging process.
References :
[1] Yuanyuan Li and Trygve O Tollefsbol. Age-related epigenetic drift and phenotypic plasticity loss: implications in prevention of age-related human diseases. Epigenomics (2016) 8(12), 1637–1651.
[2] Steve Horvath. DNA methylation age of human tissues and cell types. Genome Biology, 14:R115.
[3] Andrey Tvardovskiy, Veit Schwammle, Stefan J. Kempf, Adelina Rogowska-Wrzesinska and Ole N. Jensen. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Research, 2017, Vol. 45, No. 16, 9272–9289.
[4] Eric L. Greer and Yang Shi. Histone methylation: a dynamic mark in health, disease and inheritance, Nature Reviews, Genetics, Vol. 13 (May 2012), 343-357.
[5] Brenna S. McCauley and Weiwei Dang, Histone methylation and aging: Lessons learned from model systems, Biochim Biophys Acta. 2014 December ; 1839(12): 1454–1462. doi:10.1016/j.bbagrm.2014.05.008.
[6] Pal and Tyler Sci. Adv. 2016; 2 : e1600584
[7] Oriana Lo Re and Manlio Vinciguerra, Histone MacroH2A1: A Chromatin Point of Intersection between Fasting, Senescence and Cellular Regeneration, Genes 2017, 8, 367; doi:10.3390/genes8120367.
[8] Jia Liu, Xianhui Yin, Baohua Liu, Huiling Zheng, Guangqian Zhou, Liyun Gong, Meng Li, Xueqin Li, Youya Wang, Jingyi Hu, Vaidehi Krishnan, Zhongjun Zhou & Zimei Wang (2014) HP1α mediates defective heterochromatin repair and accelerates senescence in Zmpste24-deficient cells, Cell Cycle, 13:8, 1237-1247, DOI: 10.4161/cc.28105
[9] Jeannine Basta and Michael Rauchman. The Nucleosome Remodeling and Deacetylase (NuRD) Complex in Development and Disease, Transl Res. 2015 January ; 165(1): 36–47. doi:10.1016/j.trsl.2014.05.003.
[10] Wei Wang, Jiang-Jiang Qin, Sukesh Voruganti, Subhasree Nag, Jianwei Zhou and Ruiwen Zhang. Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications, Med Res Rev. 2015 November ; 35(6): 1220–1267. doi:10.1002/med.21358.
[11] Neil P. Blackledge, Nathan R. Rose, and Robert J. Klose. Targeting polycomb systems to regulate gene expression: modifications to a complex story, Nat Rev Mol Cell Biol . 2015 November ; 16(11): 643–649. doi:10.1038/nrm4067.
Anne Fischer

Author
Auteur
Anne is studying medicine science at the Institute of Pharmaceutical and Biological Science in Lyon and she has graduated with a Bachelor’s degree in molecular and cellular biology at the University of Strasbourg.
More about the Long Long Life team
Anne étudie les sciences du médicament à l’Institut des Sciences Pharmaceutiques et Biologiques de Lyon. Elle est titulaire d’une licence en biologie moléculaire et cellulaire de l’Université de Strasbourg.
En savoir plus sur l’équipe de Long Long Life
Dr Guilhem Velvé Casquillas

Author/Reviewer
Auteur/Relecteur
Physics PhD, CEO NBIC Valley, CEO Long Long Life, CEO Elvesys Microfluidic Innovation Center
More about the Long Long Life team
Docteur en physique, CEO NBIC Valley, CEO Long Long Life, CEO Elvesys Microfluidic Innovation Center
En savoir plus sur l’équipe de Long Long Life